Transposable Elements Can Be Utilized for Genetic Research in Drosophila
By Adam Diehl, Alan Boyle, and Elisabeth Paymal, Eye for RNA Biomedicine
Until recently, little was known about how transposable elements contribute to gene regulation. These are trivial pieces of Deoxyribonucleic acid that tin replicate themselves and spread out in the genome. Although they make up almost one-half of the man genome, these were often ignored and commonly thought of as "useless junk," with a minimal role, if any at all, in the activity of a cell. A new study by Adam Diehl, Ningxin Ouyang, and Alan Boyle, Academy of Michigan Medical School and members of the U-M Center for RNA Biomedicine, shows that transposable elements play an of import role in regulating genetic expression with implications to advance the agreement of genetic evolution.
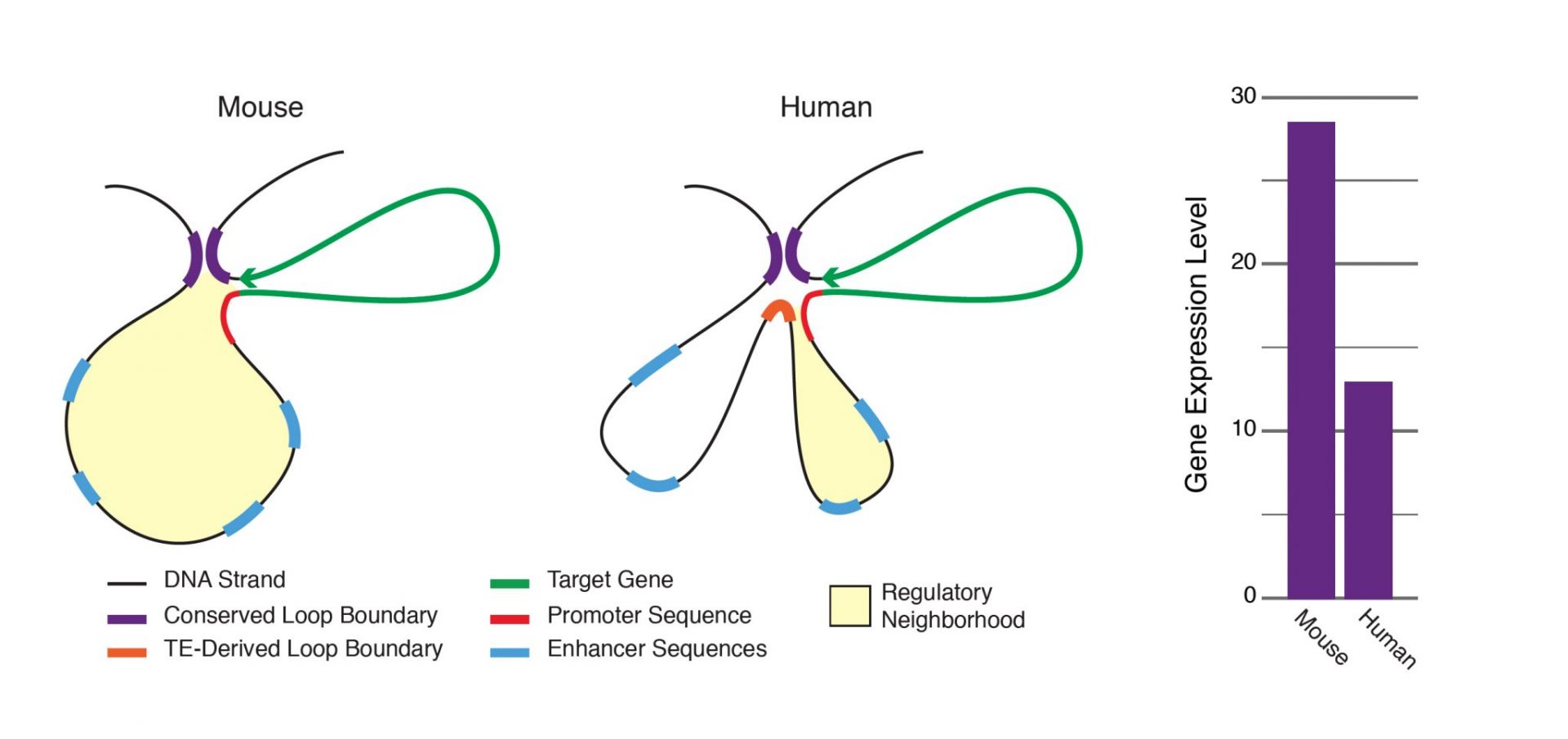
Chromatin loops are important for gene regulation because they define a gene'due south regulatory neighborhood, which contains the promoter and enhancer sequences responsible for determining its expression level. Remarkably, transposable elements (TEs) are responsible for creating around one/3 of all loop boundaries in the human and mouse genomes, and contribute upward to 75% of loops unique to either species. When a TE creates a human-specific or mouse-specific loop information technology tin change a factor's regulatory neighborhood, leading to altered factor expression. The illustration shows a hypothetical region of the human and mouse genomes in which four enhancer sequences for the aforementioned target gene fall within a conserved loop. In this case, a TE-derived loop boundary in the homo genome (orange bar) shrinks the regulatory neighborhood, preventing 2 of four enhancers from interacting with their target gene'due south promoter sequence. The internet consequence is reduced factor expression in human relative to mouse. Looping variations such equally these appear to be an important underlying cause of differential gene regulation across species and between different human cell types, suggesting that TE activity may play significant roles in evolution and illness.
Transposable elements move effectually the cell, and, unlike previously idea, the authors of this paper found that when they go to different sites, transposable elements sometimes change the way DNA strands collaborate in 3D space, and therefore the structure of the 3D genome. It appears a 3rd of the 3D contacts in the genome really originate from transposable elements leading to an outsized contribution by these regions to looping variation and demonstrating their very pregnant function in genetic expression and evolution.
The principal component that determines 3D structure is a poly peptide called CTCF. This study specially focused on how transposable elements create new CTCF sites that, in turn, hijack existing genomic structure to form new 3D contacts in the genome. The authors show that these often create variable loops that can influence regulatory activity and gene expression in the prison cell. These findings were observed in homo cells and mouse cells and evidence how transposable elements contribute to intraspecies variation and interspecies divergence, and volition guide farther research efforts in areas including gene regulation, regulatory evolution, looping divergence, and transposable element biology.
To streamline this piece of work, the authors developed a piece of software, MapGL, to track the physical proceeds and loss of short genetic sequences across species. For example, a sequence that existed in the most common antecedent may have been lost somewhere or, inversely, could have been absent in the common ancestor merely later gained in the homo genome. MapGL enables predictions almost the evolutionary influences of structural variations between species and makes this type of analysis much more attainable. For this newspaper, their input was a set of CTCF binding sites which were labeled by MapGL to show that a sequence gain/loss process explains many of the differences in CTCF binding between humans and mice.
With a groundwork in computer science and molecular biology, Alan Boyle explains that he has always been interested in gene regulation. "It'due south like a complex circuit: perturbing factor regulation through changes to the iii-dimensional structure of the genome can accept very different and broad-ranging outcomes."
For Adam Diehl, this research continues the corking discoveries that started in the late 1800s, when scientists kickoff looked at the shape of chromosomes through microscopes. They observed the shape differences between cells, and noticed that the shape within the nuclei remained the same betwixt female parent and daughter cells. Decades later on, transposable elements were discovered at his alma mater, Cornell University: jumping genes could modify the phenotypes of corn plants. In the 70s, because the genes betwixt humans and chimpanzees are much too like to explicate the differences between the species, scientific focus shifted on how the genes are being used. For Diehl "It's and then exciting to be able to synthesize all this knowledge, and contribute to the adjacent pace of the story of species development."
This inquiry team volition farther study the impact of transposable elements on the 3D genome, but this time with a particular interest on a single human population sample rather than beyond species. The side by side steps volition include experimental follow-up using a new sequencing method capable of identifying transposable element insertions that are variable across human being populations. This method was developed in collaboration with Ryan Mills's lab, at the Academy of Michigan, Medical Schoolhouse. Information technology is expected that the next results volition farther the understanding of the regulatory role of the transposable elements with possible applications to neurodegenerative diseases.
Commendation
Diehl, A.G., Ouyang, N. & Boyle, A.P. Transposable elements contribute to cell and species-specific chromatin looping and cistron regulation in mammalian genomes.Nat Commun11,1796 (2020). https://doi.org/ten.1038/s41467-020-15520-5
About the Authors
Adam Diehl is Research Calculator Specialist in the Alan Boyle Lab, Section of Computational Medicine and Biology, University of Michigan
Alan P. Boyle, Ph.D. is Banana Professor, Department of Computational Medicine and Bioinformatics (DCM&B), Department of Human being Genetics, Academy of Michigan Medical School. Alan Boyle Lab
Ningxin Ouyang is a Doctoral Pupil, Bioinformatics Candidate in the Alan Boyle Lab
Source: https://rna.umich.edu/transposable-elements/
0 Response to "Transposable Elements Can Be Utilized for Genetic Research in Drosophila"
Post a Comment